🙋🏼About Me
Hello! I am Pinglu Zhang (张平路), Phd at the  Institute of Fundamental and Frontier Sciences (IFFS), University of Electronic Science and Technology of China (UESTC) (电子科技大学,基础与前沿研究院), majoring in Computer Science and Technology.
Institute of Fundamental and Frontier Sciences (IFFS), University of Electronic Science and Technology of China (UESTC) (电子科技大学,基础与前沿研究院), majoring in Computer Science and Technology.
I am also part of a joint training program (联合培养) at the  Zhongguancun Academy(北京中关村学院).
Zhongguancun Academy(北京中关村学院).
I am conducting research on Sequence Alignment at the Malab laboratory under the supervision of Prof. Quan Zou (邹权教授). My work focuses on developing multiple sequence alignment for large-scale data, mulitiple genome alignment, centromere region alignment, and related topics 
Researchers interested in collaboration are welcome to contact me at pingluzhang@outlook.com.
🔥 News
- 2025.12.11: 🎉🎉 NSFC Young Student Basic Research Grant (国自然博士生项目)!
- 2025.11.30: 🎉🎉 New article has been accepted by Genome Biology!
- 2025.02.07: 🎉🎉 New article has been accepted by Genome Research!
- 2024.12.30: 🎉🎉 I have been offered a joint PhD position at UESTC and Zhongguancun Academy.
- 2024.09.08: 🎉🎉 New Homepage was released!
💰 Fundings
- 国家自然科学基金青年学生基础研究项目(博士研究生) 面向全基因组的大规模多物种基因组比对研究
📝 Publications
Selected Publication
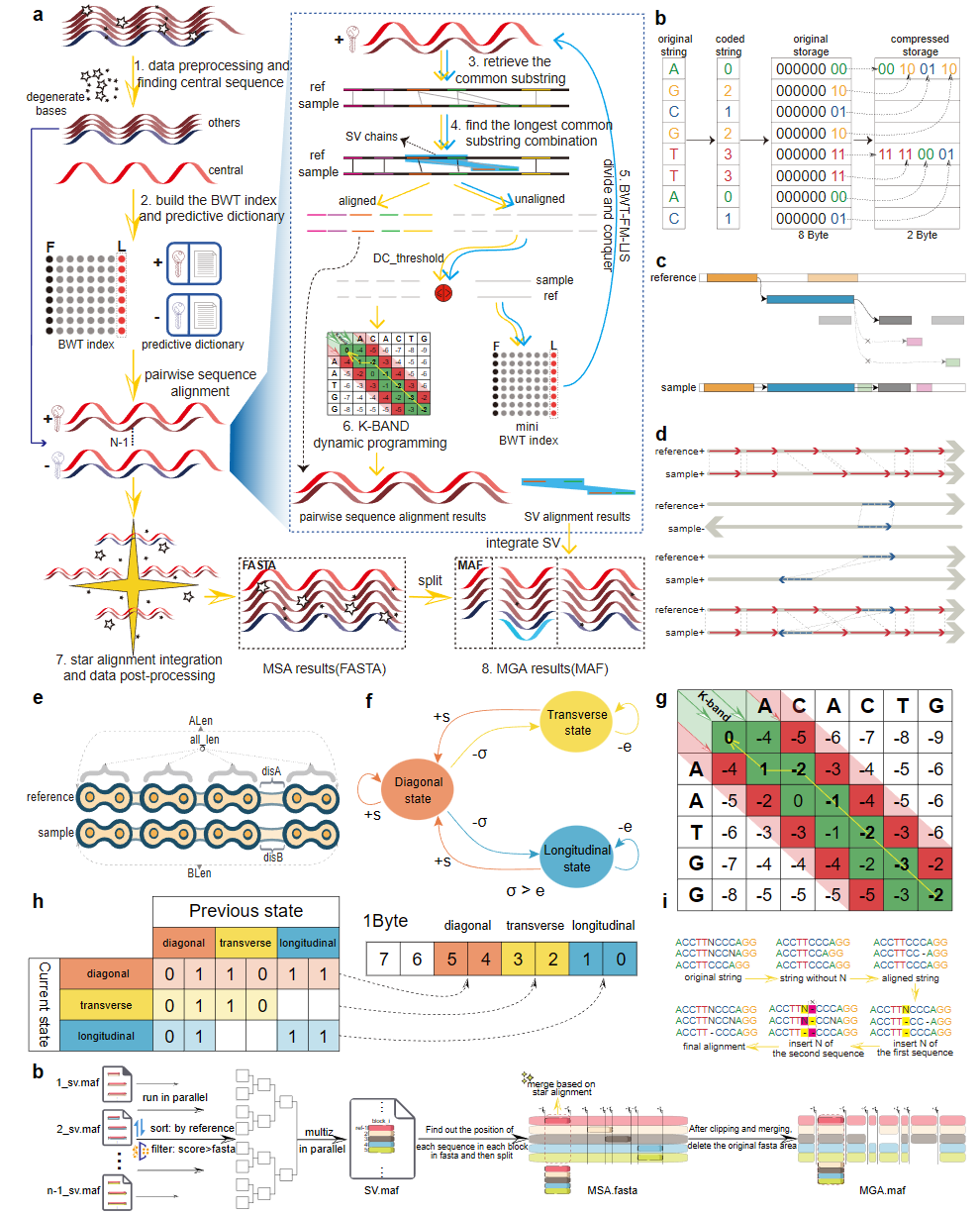
HAlign-G: rapid and low-memory multiple-genome aligner for large-scale closely related genomes
Pinglu Zhang†, Tong Zhou†, Yanming Wei, Qinzhong Tian, Yixiao Zhai, Yizheng Wang, Quan Zou, Furong Tang*, Ximei Luo*
Genome Biology, 2025, 中科院1区, IF2025=9.4
-
HAlign-G is a fast and memory-efficient solution for large-scale multiple genome alignment, integrating BWT-FM-LIS indexing, an optimized K-band algorithm, and a star-alignment strategy.
-
It includes HAlign-G1 for intra-species multiple sequence alignment and HAlign-G2 for cross-species multiple genome alignment, each optimized for distinct evolutionary signals.
-
Benchmarks indicate that HAlign-G achieves higher accuracy, lower memory usage, and significantly faster performance than existing methods across both short- and long-genome datasets.
Fast sequence alignment for centromere with RaMA
Pinglu Zhang, Yanming Wei, Qinzhong Tian, Quan Zou, Yansu Wang*
Genome Research, 2025, 中科院1区, nature index journal, IF2024=6.24.
- RaMA is a novel sequence alignment tool designed for centromeric regions, leveraging rare matches as anchors and a 2-piece affine gap cost to capture genetic evolution accurately.
- RaMA utilizes parallel computing and the wavefront algorithm, achieving up to 13.66 times faster processing and using only 11% of UniAligner’s memory.
- Additionally, RaMA introduces two innovative methods for defining reliable alignment regions, which further refine the accuracy of centromeric alignment statistics and provide more robust insights into genetic variations.
- Downstream analysis of simulated data and HOR arrays demonstrates RaMA’s superior accuracy in capturing true HOR structures and defining reliable alignment regions.
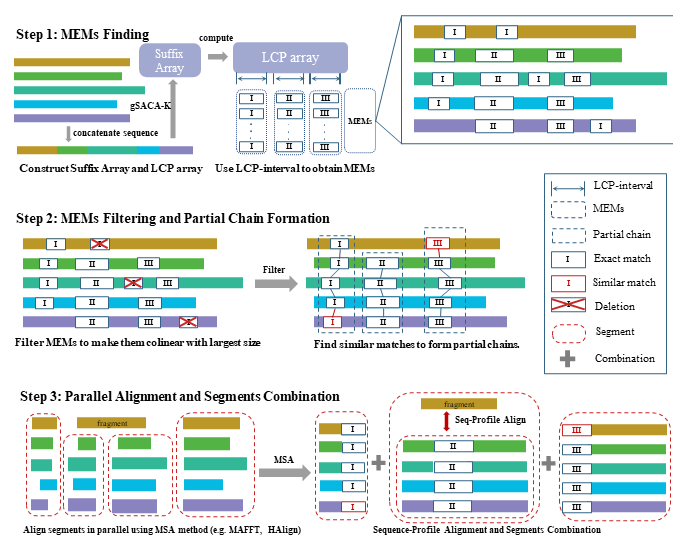
FMAlign2: a novel fast multiple nucleotide sequence alignment method for ultralong datasets
Pinglu Zhang, Huan Liu, Yanming Wei, Yixiao Zhai, Qinzhong Tian, Quan Zou*
Bioinformatics, 2024, 中科院3区, CCF-B, IF2024=4.4.
- FMAlign2 is an improved MSA method that uses a suffix array and vertical division strategy to align ultralong sequences in parallel.
- FMAlign2 reduces processing time while maintaining accuracy, handling sequences up to billions in length efficiently.
Other Publications
-
PS-mixer: A polar-vector and strength-vector mixer model for multimodal sentiment analysis, H Lin†, P Zhang†, J Ling, Z Yang*, LK Lee, W Liu. Information Processing & Management, 2023, 中科院1区, CCF-B, IF2023=8.6. Paper Code
-
Chimera: Ultrafast and Memory-efficient Database Construction for High-Accuracy Taxonomic Classification in the Age of Expanding Genomic Data, Q Tian†, P Zhang†, Y Wei, Q Zou, Y Wang*, X Luo*. bioRxiv, 2025. Paper Code
-
HAlign-4: A New Strategy for Rapidly Aligning Millions of Sequences, T Zhou, P Zhang, Q Zou*, W Han*. Bioinformatics, 2024, 中科院3区, CCF-B, IF2024=4.4. Paper Code
-
TPMA: A two pointers meta-alignment tool to ensemble different multiple nucleic acid sequence alignments, Y Zhai, J Chao, Y Wang, P Zhang, F Tang*, Q Zou*. PLOS Computational Biology, 2024, 中科院2区, CCF-B, IF2024=3.8. Paper Code
-
ReAlign-P: A vertical iterative realignment method for protein multiple sequence alignment, Y Zhai, P Zhang, Q Zou, X Luo*. Bioinformatics, 2025, 中科院3区, CCF-B, IF2025=5.4. Paper Code
-
Application and Comparison of Machine Learning and Database-Based Methods in Taxonomic Classification of High-Throughput Sequencing Data, Q Tian, P Zhang, Y Zhai, Y Wang*, Q Zou*. Genome Biology and Evolution, 2024, 中科院2区, IF2024=3.2. Paper Code
-
ReAlign-Star: An Optimized Realignment Method for Multiple Sequence Alignment, Targeting Star Algorithm Tools, Y Zhai, P Zhang, Y Liu, Q Zou*. International Conference on Intelligent Computing, 2025, CCF-C. Paper Code
-
TCM@MPXV: A Resource for Treating Monkeypox Patients in Traditional Chinese Medicine, Xin Zhang, Feiran Zhou, Pinglu Zhang, Quan Zou* and Ying Zhang*. Current Bioinformatics, 2024, 中科院3区, IF2024=2.4. Paper
🎖 Awards
- 2023.09 The First Prize (Ranked 1st Overall) in the CBC Data Challenge (CBC数据挑战赛全国一等奖).
- 2025.03 电子科技大学优秀研究生和学术青苗.
📖 Educations
-
2023.09 - (now): Phd, Institute of Fundamental and Frontier Sciences (IFFS), University of Electronic Science and Technology of China (UESTC) (电子科技大学,基础与前沿研究院).

-
2025.03 - (now): Phd (Joint Program), Zhongguancun Academy(北京中关村学院).

-
2024.06 - 2025.03: Master (Joint Program), Yangtze Delta Region Institute (Quzhou), University of Electronic Science and Technology of China (UESTC) (电子科技大学,长三角研究院).

-
2019.09 - 2023.06: Bachelor, School of Computer Science, Guangdong University of Technology (GDUT) (广东工业大学,计算机学院).

